CML paper summary - January 2025
The e13a3 (b2a3) and e14a3 (b3a3) BCR::ABL1 isoforms are resistant to asciminib
Leske IB and Hantschel O. Leukemia, September 2024
Leukemia 2024 Sep;38(9):2041-2045: https://www.nature.com/articles/s41375-024-02314-7
Introduction:
Asciminib, a first-in-class allosteric BCR::ABL1 inhibitor, has shown significant efficacy in patients with Philadelphia chromosome-positive chronic myeloid leukemia (CML) who have been previously treated with other tyrosine kinase inhibitors (TKIs). This study by Leske and Hantschel investigates the potential resistance of specific BCR::ABL1 isoforms to asciminib, particularly focusing on the e13a3 (b2a3) and e14a3 (b3a3) isoforms. These isoforms, which lack exon 2 of ABL1 and are present in less than 1% of CML patients, hypothesised to be resistant to asciminib due to the absence of a functional SH3 domain crucial for the drug's mechanism of action.
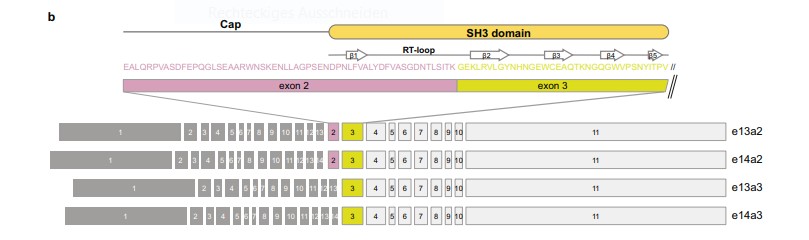
Fig 1: Overview of BCR::ABL1 exon organisation of the common e13a2, e14a2 transcript variants, as well as e13a3 and e14a3 that lack ABL1 exon 2.
Study design:
- Objective: To evaluate the resistance of BCR::ABL1 e13a3 and e14a3 isoforms to asciminib treatment.
- Methodology: The study involved generating cDNAs for the e13a3 and e14a3 isoforms and testing their response to asciminib in BaF3 cells. The effect of asciminib on BCR::ABL1-dependent cell proliferation and survival was measured using a range of asciminib concentrations.
- Comparison: The results for e13a3 and e14a3 were compared to the more common e14a2 isoform, which is known to be sensitive to asciminib.
Study results:
- Asciminib sensitivity:
- The e13a3 and e14a3 isoforms demonstrated significant resistance (~10,000-fold) to asciminib, with no inhibition observed at concentrations below 50 µM. In contrast, the e14a2 isoform showed a half-maximal growth inhibition (GC50) of approximately 1 nM.
- Both e13a3 and e14a3 isoforms were highly sensitive to dasatinib, an ATP-competitive inhibitor.
- Mechanism of resistance:
- The resistance observed for the e13a3 and e14a3 isoforms was attributed to the absence of a functional SH3 domain, which is required for asciminib's allosteric inhibition of BCR::ABL1. Structural modelling and binding studies confirmed that the lack of exon 2 disrupts the SH3 domain, preventing the necessary conformational changes for asciminib-mediated inhibition.
- Immunoblotting revealed that STAT5A/B phosphorylation, critical for leukemogenesis, remained unaffected by asciminib in e13a3 and e14a3 cells.
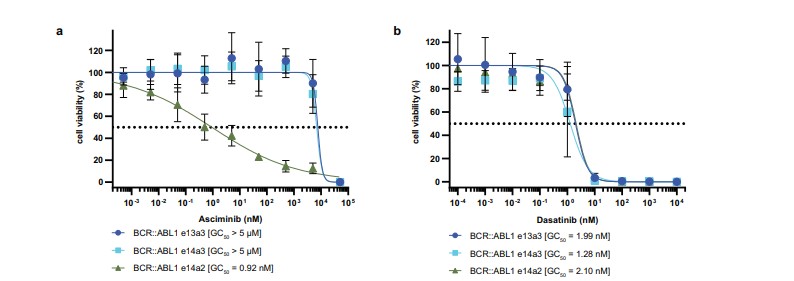
Fig 2: BCR::ABL1 e14a2 and e14a3 are asciminib resistant, but sensitive to dasatinib. Normalised viability of Ba/F3 cells expressing BCR::ABL1 e14a2, e14a3 and e13a3 fusion transcripts was measured in the presence of the indicated concentrations of asciminib (a)
or dasatinib (b) after 48 h.
Conclusion
This study provides evidence of a primary resistance mechanism to asciminib in CML patients with the e13a3 and e14a3 BCR::ABL1 isoforms. These findings are crucial for personalising treatment strategies in CML and suggest that ATP-competitive BCR::ABL1 inhibitors, such as dasatinib, should be preferred for patients with these isoforms. Genetic screening for BCR::ABL1 isoforms is crucial before initiating therapy to ensure optimal treatment selection for CML patients.










