The iCMLf Genomics Alliance - Investigating clonal hierarchy in CML
The iCMLf Genomics Alliance - Investigating clonal hierarchy in CML
The iCMLf/HARMONY Genomics project is analysing genetic data from hundreds of CML patients, aiming to uncover the hierarchy of genomic events in CML. This will enhance our understanding of how genetic variations impact treatment outcomes.
Questions we hope to answer include:
- Clinical relevance of mutations at diagnosis?
- Do some mutations confer a poorer prognosis?
- Can poor risk be overcome with more potent drugs?
- Will mutations at diagnosis impact the chance of treatment-free remission?
Progress to date:
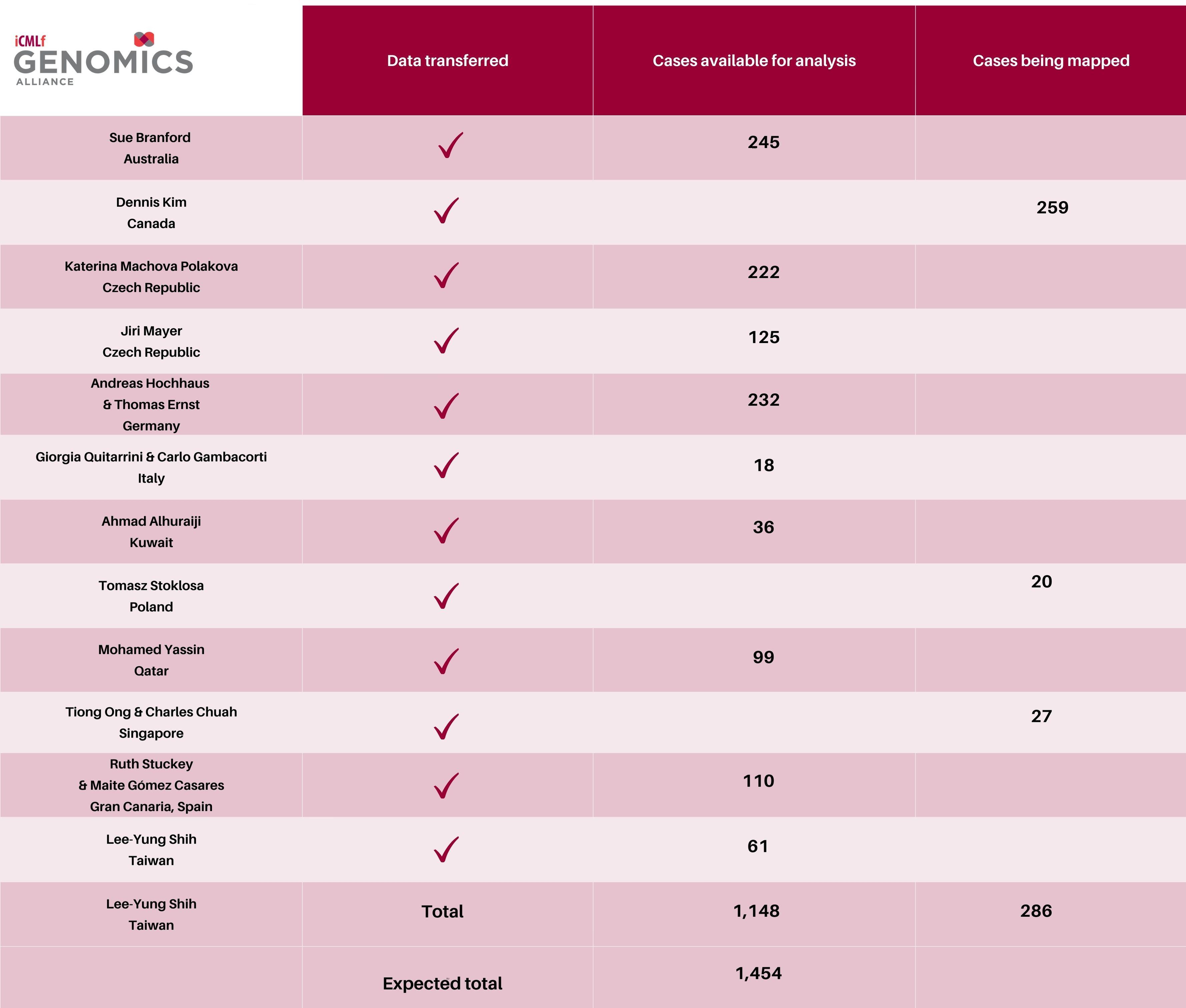
- All sites now completed data upload!
- 9 sites finalised, with upload, transfer and mapping completed
- 3 sites finalising mapping
- 1,148 cases available for analysis with approximately 300 still coming.
Data Analysis is Underway
Data analysis has begun on 1,044 cases submitted with NGS data.
- Work performed so far:
- Revision and re-harmonisation of 760 variants
- Work in Progress
- Extraction of non-mutated cases
- Identification of disease phases at NGS available dates
We welcome further interest from groups to contribute data, or ideas for future analysis.
We thank Incyte and Pfizer for supporting the iCMLf Genomics and TFR Alliances.
In partnership with the HARMONY Foundation Big Data platform
History of the iCMLf/HARMONY research project to investigate clonal hierarchy in CML
In 2008 the iCMLf established a global CML Genomics Alliance to answer key clinical questions around the role of genetic mutations in clinical outcomes for CML. A literature review conducted by the iCMLf CML Genomics Alliance1, examined studies that reported mutated genes in adult patients with CML and established a clear, clinical need for further genomic evaluation in CML.
Many clinical questions could be answered by pooling data. For example:
- Clinical relevance of mutations at diagnosis?
- Do some mutations confer a poorer prognosis?
- Can poor risk be overcome with more potent drugs?
- Will mutations at diagnosis impact the chance of treatment-free remission?
CML is characterised by genomic aberration of the BCR-ABL1 gene, but other cancer gene mutations are detected at diagnosis and in advanced phases and may be more frequent than the BCR-ABL1 kinase domain mutations.
The iCMLf Genomics Alliance research project aims to elucidate the hierarchy of genomic events that underlie the disease course in CML. Key research questions are to investigate if certain mutations might be associated with an increased risk in terms of TKI response and progression and how this knowledge could influence our therapeutic decision-making. The ultimate goal is to improve CML management and patient outcomes.
To obtain insights into the role of mutations other than BCR-ABL1 kinase domain mutations, a large database with clinical and genetic data from thousands of patients is required.
HARMONY Plus platform – Big data to improve CML treatment
‘The HARMONY Plus Big Data platform now provides the opportunity to move forward to answer multiple clinical questions in CML’ (Sue Branford)
The HARMONY Big Data Project facilitates data pooling and sharing for specific hematologic malignancies with a central repository of data. Genomic and clinical data is collected from multiple research groups that contribute anonymized data in a secure way following all legal and ethical requirements.
HARMONY Plus is an extension to the project to expand the scope of data sharing to other hematologic malignancies, including CML. The HARMONY Plus project team is collaborating with the iCMLf Genomic Alliance to facilitate the contribution of clinical and genetic CML data to HARMONY Plus to answer key clinical questions using Big Data.
iCMLf invites the CML community to contribute data
The iCMLf encourages partners and other interested parties to participate in the project, to define variables for data collation and to commence the submission of datasets to HARMONY Plus.
If you are interested to contribute data or ideas to HARMONY Plus, please email info@cml-foundation.org.
Make a difference in the fight against CML
You can donate to the iCMLf research projects by visiting our donation page to get involved and make a difference in the fight against CML.
We are grateful to Pfizer and Incyte for support for the analysis and publication of the iCMLf Alliance projects.
(1) Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Branford S et al. Leukemia 2019:33:1835